What
What can STRmix™ do?
With an ever growing range of functions underpinned by world leading statistical and biological modelling, STRmix™ enables forensic biologists to:
Resolve mixed DNA profiles without reference to known contributors.
Enter a contributor number range when performing a deconvolution.
Assign an LR varying the number of contributors under the prosecution and defence propositions.
Undertake quality checks for data.
Set the number of major contributors to a mixed DNA profile you are interested in and obtain an LR only for these.
Model any type of stutter observed within your STR profiling kit, now including improved dropout modelling allowing analysis of DNA profiles with a low, or even no analytical threshold.
Compare reference DNA profiles to single source and mixed DNA profiles and provide a statistical weighting.
Interpret DNA profiling data generated by any autosomal STR profiling kit, now including Amelogenin in the interpretation.
Interpret DNA profiles from a range of starting template DNA concentrations.
Use laboratory-specific settings to perform calculations suited exactly to that laboratory’s results.
Set up multiple report templates and generate more than one report at the end of each calculation.
Visualise a representation of the evidence input and ignore any peak during setup.
Review the deconvolution and explore the accepted genotypes, their weights, and how a person of interest aligns.
Search a deconvoluted DNA profile directly against a database without the need to interpret a single source component.
Collate your deconvolution and likelihood ratio data using the in built tools.
Calculate multiple LRs from multiple reference inputs to a previously run deconvolution (LR Batch tool).
Perform a large number of in-silico specificity tests on a profile-by-profile basis (Hd True Tester tool).
Batch multiple deconvolutions or other STRmix™ functions (such as Interpretation, LR from Previous, and Database Search) in a queue, allowing the user to run multiple deconvolutions and calculate LRs sequentially.
Instantly set up interpretations (including replicate inputs) with flexible likelihood ratio propositions for multiple profiles using Batch Maker.
Combine multiple amplifications of the same DNA extract into one interpretation, even when generated with different multiplexes.
Accommodate data generated by protocols demonstrating increased stochastic variation and non-zero allelic drop-in rates, for example elevated PCR cycle number and enhanced CE injection methods.
Include related individuals as alternate propositions in the LR.
Carry out familial searches against a database, searching for close relatives of contributors to mixed DNA profiles.
Optionally utilise peak labelling probabilities in STRmix™ deconvolutions (An advanced feature for FaSTR™ DNA users).
Generate fully configurable (and if required, retrospective) reports including a CODIS report.
Password protect default settings and kit settings.
STRmix™ IN USE
STRmix™ has been used for routine casework interpretation at PHF Science and FSSA since August 2012 and is now the Australasian standard for DNA interpretation.
In one case, a mixed DNA profile was obtained from a semen stain on a carpet at the scene of an alleged sexual assault involving two male offenders. DNA present in approximately equal proportions from two individuals was detected (see EPG below). This profile was unsuitable for database searching using traditional DNA interpretation methods. Using STRmix™, the profile was interpreted assuming the profile originated from two contributors and compared against a database with over 145,000 profiles. The crime profile matched two individuals.
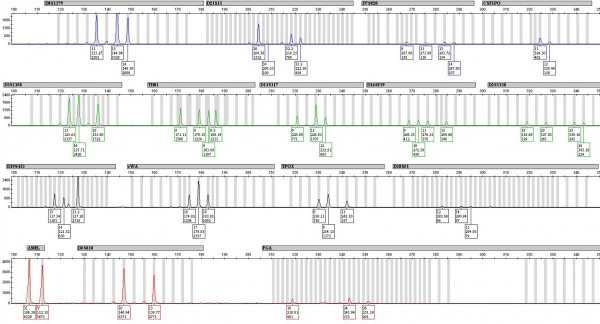
This case highlights how STRmix™'s database search function can provide investigative information. It can also be used to validate complex mixtures where contamination is suspected.
